
Differentiable dynamic programming for structured prediction and attention. Estimating amino acid substitution models: a comparison of dayhoff’s estimator, the resolvent approach and a maximum likelihood method. TensorFlow: large-scale machine learning on heterogeneous systems (, 2015) Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. et al.) 4171–4186 (Association for Computational Linguistics, 2019). 1 (Long and Short Papers) (eds Burstein, J. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2019 Vol. BERT: pre-training of deep bidirectional transformers for language understanding. Pfam: the protein families database in 2021. Uniref clusters: a comprehensive and scalable alternative for improving sequence similarity searches.

of the 31st International Conference on Neural Information Processing Systems (eds Guyon, I. End-to-end learning of multiple sequence alignments with differentiable Smith-Waterman. Protein structural alignments from sequence. In 7th International Conference on Learning Representations (ICLR) (, 2019). Learning protein sequence embeddings using information from structure. In Research in Computational Molecular Biology (eds Apostolico, A. Optimizing amino acid substitution matrices with a local alignment kernel. In Computing and Combinatorics (eds Ibarra, O. Pfasum: a substitution matrix from pfam structural alignments. Amino acid substitution matrices from protein blocks.
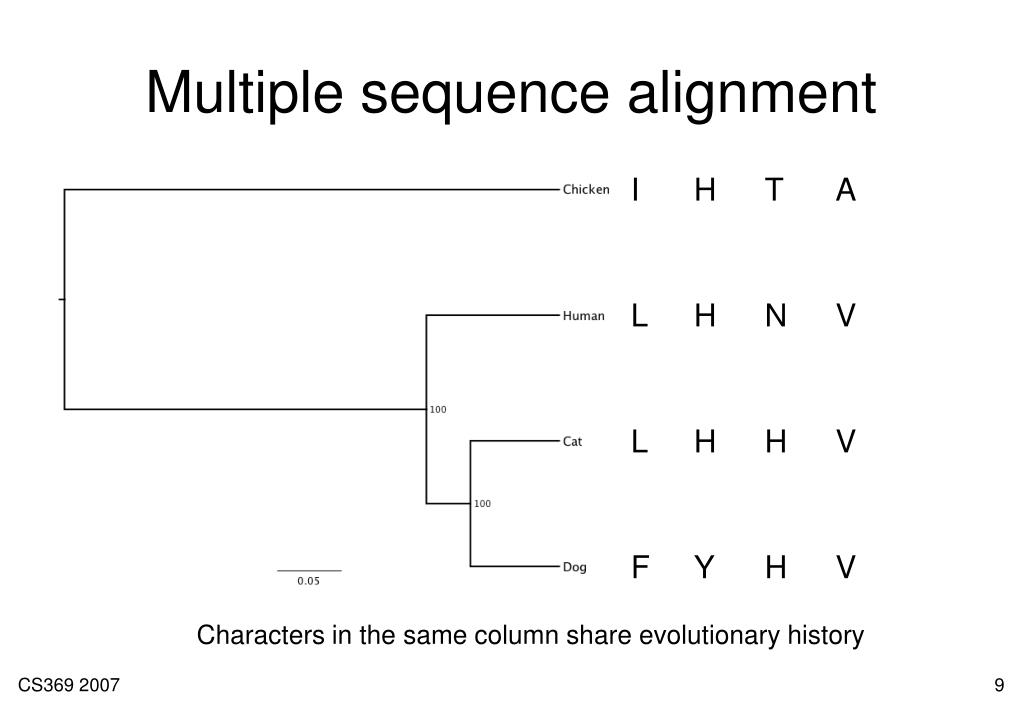
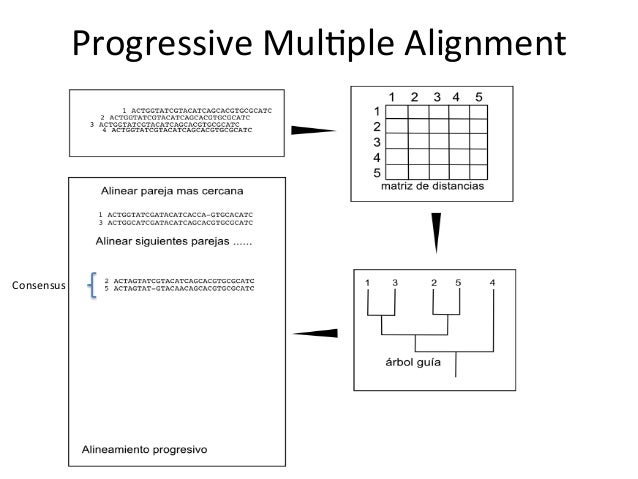
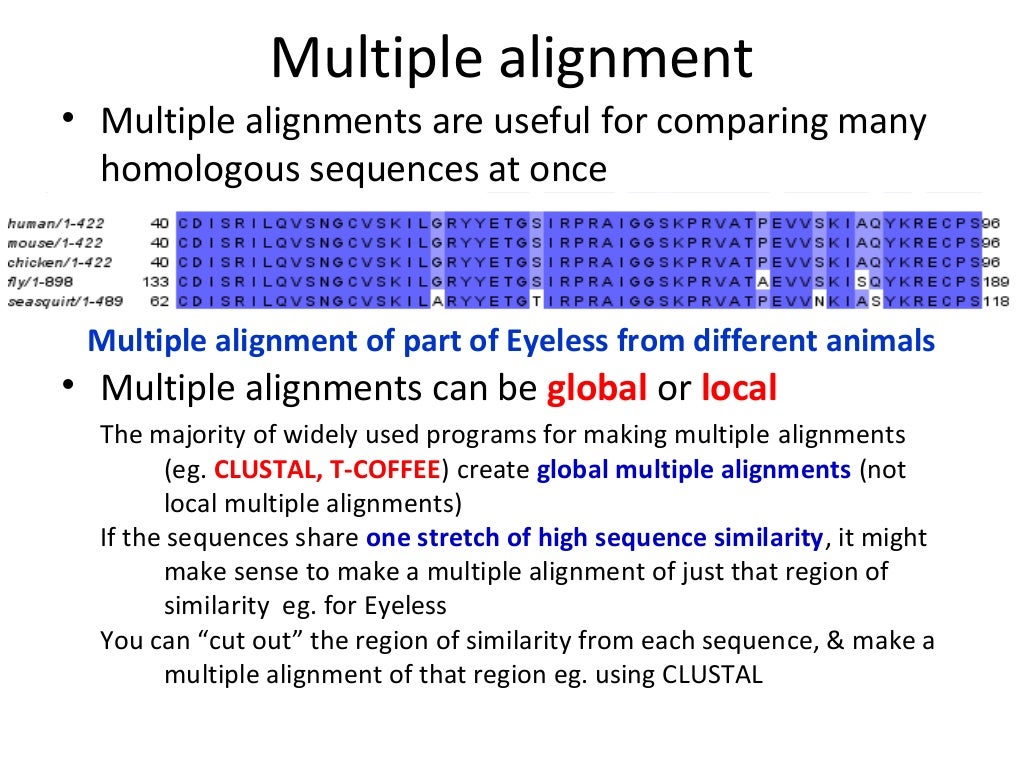
Parametric inference for biological sequence analysis. Parametric and inverse-parametric sequence alignment with xparal. review of concepts, case studies and implications. Parametric and ensemble sequence alignment algorithms. Parametric optimization of sequence alignment. Support vector training of protein alignment models. Remote homology and the functions of metagenomic dark matter. Characterization of pairwise and multiple sequence alignment errors. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Rapid and sensitive sequence comparisons with FASTP and FASTA. Identification of common molecular subsequences. Highly accurate protein structure prediction with alphafold. Protein 3D structure computed from evolutionary sequence variation. Evolutionarily conserved pathways of energetic connectivity in protein families. Functional assignment of metagenomic data: challenges and applications. Once trained, we show that DEDAL improves by up to two- or threefold the alignment correctness over existing methods on remote homologs and better discriminates remote homologs from evolutionarily unrelated sequences, paving the way to improvements on many downstream tasks relying on sequence alignment in structural and functional genomics. DEDAL is a machine learning-based model that learns to align sequences by observing large datasets of raw protein sequences and of correct alignments. Here we leverage recent advances in deep learning for language modeling and differentiable programming to propose DEDAL (deep embedding and differentiable alignment), a flexible model to align protein sequences and detect homologs. Aligning highly divergent sequences remains, however, a difficult task that current algorithms often fail to perform accurately, leaving many proteins or open reading frames poorly annotated. Protein sequence alignment is a key component of most bioinformatics pipelines to study the structures and functions of proteins.


 0 kommentar(er)
0 kommentar(er)
